Technical report
Optimal breeding strategy for mouse mutant strainss
by Gaël Grannec, Marika Nosten-Bertrand
 Correspondence: Institut du Fer à Moulin
Correspondence: Institut du Fer à Moulin
INSERM UMR-S 839 UPMC
17 rue du Fer à Moulin
75005 Paris, France
e-mail: gael.grannec@inserm.fr; marika.nosten-bertrand@inserm.fr
Tel : 33 (0) 1 45 87 61 43
Fax : 33 (0) 1 45 87 61 32
Summary
Despite the published recommendations after the Banbury Conference in 1997 to maintain a mutation on controlled genetic backgrounds, maintenance of mutant strains and production of experimental groups are still too often hampered by constraints of time, space, and cost.
We propose here a simple and rigorous method that allows not only the efficient production of animals, but especially the precise control of the genetic background and the generation of appropriate control groups, guaranteeing stable and reproducible observations. This method is flexible and allows optimization and adaptation of the production according to the experimental needs, thus reducing the final cost. In addition, the work of the Animal Care technicians is simplified and animal welfare is improved.
Introduction
Whether a mutation has appeared spontaneously, or was induced pharmacologically or by means of genetic engineering, fixing it onto an inbred genetic background is a sine qua non condition not only to generate optimal control groups, but also to ensure the stability and reproducibility of results in space and time.
These theoretical and practical issues related to the impact of
genetic background on the phenotypic expression of a mutation are so
essential that the scientific community has in the past joined forces
to educate researchers, provide guidelines and encourage them to
standardize conditions to maintain their mutations of interest and
produce experimental animals (Banbury workshop report, 1997).
The first recommendation sustains that the genetic background is
described in detail and is easily reproducible. The second
recommendation prompts researchers to derive their targeted mutation
simultaneously on two standard inbred strains, as shown in Figure 1.
|
Figure 1. Maintaining a genetic mutation
simultaneously on two inbred genetic backgrounds (A and B)
allows the production of inbred and / or hybrid (AxB-F1)
experimental animals. Click image to enlarge |
When the mutation of interest is transferred by successive backcrosses
simultaneously on two inbred genetic backgrounds, heterozygotes from
each congenic strain Am and Bm are used for the production of
experimental animals: a cross between heterozygotes (HT) allows the
study of the phenotypic expression of the mutation alternately or
simultaneously on inbred (Figure 1b) and hybrid (Figure 1c) genetic
backgrounds. Given the complexity of interactions between genes
(epistasis), expression of a mutation varies depending on the genetic
environment in which it is expressed. The analysis of a mutation
simultaneously on two different genetic backgrounds is a powerful tool
to identify modifier genes that interact with the mutation and
contribute to the complexity of phenotypes (Banbury workshop report
1997; Doetschman 2009). In addition, it allows the phenotypes to be
evaluated on an F1 hybrid background that can be very favorable when
the homozygous mutation is lethal on an inbred background for example.
As shown on Figure 1a, heterozygotes are also used to maintain the
congenic strains: consistent backcrosses with identified breeders from
approved suppliers are crucial to reduce the risk of genetic drift and
thus maintain a stable genetic background along time.
The choice of the optimal background is therefore an important step to
maintain a mutation. Since the first DBA inbred strain developed by
Little in the 1900s, more than 450 inbred strains have been described,
and for the most widely used, many substrains have been derived for so
many generations that they have to be considered distinct (Simon et
al. 2013; Fontaine and Davis 2016). The choice is even increasing with
the ongoing development of new inbred strains (Srivastava et al. 2017)
providing a wealth of different genotypes and phenotypes, each having
a unique and stable genetic environment that allows phenotypic
measures on isogenic individuals (Beck et al. 2000).
How to translate these recommendations specifically in order to
organize proper breeding within animal facilities? How to reconcile
these requirements with constraints of time, space and budget that all
researchers are facing today? To answer these questions, we developed
a simple method that adapts to all types of breeding, either reduced
to maintain the mutation and produce regular breeders at low returns,
for example, or enlarged, for simultaneously generating a large number
of experimental animals required for behavioral or pharmacological
studies.
The principle:
The method, presented in Figure 2 for four cages, is to use one male
for four females (one female per cage) for periodic and controlled
coupling. The male is rotated from one cage to another every fortnight
(thus staying with each female for 3 to 4 estrous cycles). After four
moves, the male is back to the first female. Each female spends two
weeks with the male, then one week with another female for company,
preferably of a passive strain of a different coat color for easy
identification. During the fourth week, the breeding female is left
alone for parturition and lactation for four weeks. After weaning, the
breeding female recovers for one week with the female companion before
the return of the male. Therefore each female produces a litter every
eight weeks, and the yield is two litters per month for four cages.
This systematic sequence is particularly flexible and therefore
suitable for all types of desired production. We give here the most
useful variants:
- With only four cages, the production can be doubled with two breeding females per cage. The female companion is no longer necessary. Both females become quickly synchronized and ready for mating during the first days of the mating. In this option, the yield is two litters every two weeks. The presence of two females optimizes maternal environment (Sayler and Salmon 1969) and for experimental subjects, the pre-weaning environment is homogenized. In case of death of one female, the number of pups can be reduced (for example by choosing one gender) in order to save animals of interest without adoption constraints.
- To produce larger experimental groups, the number of cages can be doubled. In this option, from 8 cages of two females and two breeding males, 8 litters per month are obtained, and it is still possible to modulate this production: the two males can be rotated simultaneously for a production of four litters every two weeks, or shifted by one-week to obtain two litters per week.
- Within the same animal facility, several mutations are often maintained on the same genetic background. The same male can thus serve four cages of breeding females carrying different mutations. This option is particularly suitable to optimize tight spaces.
- Some protocols require earlier weaning (3 weeks). In this option, the male is left for one week with two breeding females. With 14 females in 7 cages, two litters can be obtained per week.
This method and its variations are suitable for autosomal as well as X-linked mutations and for both the maintenance of the mutant strains (by consistent backcrossing onto defined inbred background) and the production of experimental groups (Figure 1). For autosomal mutations, production with both breeders heterozygous for the mutation gives rise to wild-type (WT), knockout (KO) and heterozygous (HT) pups in the Mendelian’ proportions 1/4, 1/4 and 1/2, respectively. Note that the WT littermates are the optimal control group, sharing with the experimental KO and HT subjects not only the same genetic background, but also the same pre-and post-natal maternal environments. In the case of X-linked mutations, conventional backcross of a heterozygous X+/X- female with a WT male serves simultaneously the maintenance of the genetic background and the production of experimental groups of males (X+/Y and X-/Y) and females (X+/X+ and X+/X-).
|
Figure 2. Method for optimized breeding (periodic couples): Example of four cages regime. With four females (F1-F4) per
male, each female is successively in the presence of the male
for two weeks, a female companion (FC) for one week, and its
litter for four weeks before one week of rest with the
companion, and then the male again to re-start the cycle. Each
female produces a litter every eight weeks and the yield of two
litters per month for four cages can be doubled with two
breeding females per cage (in this case, the female companion is
not needed). Click image to enlarge |
In terms of husbandry procedures, this systematic sequence has many
advantages to enhance the welfare of laboratory mice. We particularly
emphasize the absence of the male during parturition and lactation,
which is essential for optimized breeding quality. In most
laboratories, mating during a postpartum oestrus is often used in
order to shorten inter litter interval and increase the yield
(Mantalenakis and Ketchel 1966). Continuous mating, however, implies
simultaneous pregnancy and lactation that increased cost for the dam:
since laboratory mice have similar gestation and lactation lengths,
the peak demand of pregnancy overlaps with the peak demand of
lactation, thus increasing the energy burden experienced (Johnson,
Thomson and Speakman, 2001). Classical mouse genetic literature has
shown that concurrent lactation was responsible for delayed
implantation, increased post-implantation mortality, and potential
morbid effects on the pups subsequently born in the second litter,
including long-lasting detrimental effects (McCarthy 1965; Eisen and
Saxton 1984; Fone and Porkess 2008; Lerch et al. 2015). For postpartum
fertilization, the duration of pregnancy is increased, and the extent
of the delay in implantation is correlated with the number of suckling
pups, which may directly impact developmental studies (Mantalenakis
and Ketchel 1966; Bindon 1969; Norris and Adams 1981). In addition,
the immune status of pregnant females during lactation is also
affected, which influences the descendants and weakens the health of
the entire colony (Lloyd 1983). In the present sequence, one week of
post-weaning “reset” for breeding females preserves their
hormonal and immune states.
Note that a few exceptional strains - wild inbred for example - may
only tolerate permanent couples. In these cases, females are not
receptive to the male throughout lactation (Guenet JL, personal
communication). But for most standard inbred strains, the absence of
the male during lactation is an essential factor that maintains
efficient litter sizes and the health quality of animal facilities and
contributes to optimal breeding.
We also insist on the fact that in such breeding sequences, animals
are never isolated, limiting stress (Valzelli 1973; van Loo et al.
2003; Varty et al. 2006; Fone and Porkess 2008). Finally, by setting
the weaning age at one month, the sequence is not disturbed even in
case of late fertilization and it adapts to most strains, including
when the mutation induces a pre-weaning developmental delay.
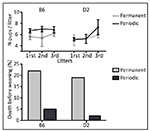
The Figure 3 presents the reproductive performance that we obtained in
our animal facilities to maintain a mutation simultaneously on two
inbred C57BL/6NCrl (B6) and DBA/2J (D2) genetic backgrounds by regular
backcrosses. This retrospective analysis shows that, breeding within
our facilities with periodic couples, compared to permanent couples,
improved the litter size at birth, but only significantly in the B6
strain (F (1,75) = 21.7, P = 0.03) and this effect was observed from
the first litter. In addition, a 4-fold reduction of pre-weaning
deaths was observed in both B6 and D2 strains.
|
Figure 3. Retrospective analysis of reproductive performance
of permanent compared to periodic couples. Click image to enlarge |
It is worth noting that this systematic sequence has also advantages
for the animal care staff. Periodic couples facilitate the monitoring
of the breeding colonies, with programmed production and constant
rates. The work of the Animal Care technicians is simplified: a weekly
visit on a fixed day is enough to handle the breeding program, which
consists successively of changing the company female, the male, or
weaning the litters.
In conclusion, this method provides birth control that reduces the
number of animals produced, with a higher survival rate, in fitting
with the 3Rs (reduce, refine, replace), established principles in
operation worldwide (Richmond 2000).
Acknowledgements
The authors gratefully acknowledge Christine Lamouroux (ex head of the Murine Platform at the Institute of Integrative Biology, University Pierre et Marie Curie, Paris) and her team; Bruno Giros for his confidence and Fiona Francis and Melissa Stouffer for her helpful reading of this manuscript.
References
- Banbury Workshop Report, (1997). Mutant mice and neuroscience: recommendations concerninggenetic background. Banbury Conference on genetic background in mice. Neuron. 19, 755-759.
-
Beck, J.A., Lloyd, S., Hafezparast, M., Lennon-pierce, M., Eppig,
J.T., Festing, M. F., Fisher, E. M., (2000). Genealogies of mouse
inbred strains. Nature Genetics. 24,
23-25.
Bindon, B.M., (1969). Mechanism of the inhibition of implantation in suckling mice. Journal of Endocrinology. 44, 357-362. - Doetschman, T., (2009). Influence of genetic background on genetically engineered mouse phenotypes. Methods in Molecular Biology. 530, 423-433.
- Eisen, E.J., Saxton, A.M., (1984). Effects of concurrent lactation and postpartum mating on reproductive performance in mice selected for large litter size. Journal of Animal Science. 59, 1224-1238.
- Fone, K.C., Porkess, M.V., (2008). Behavioural and neurochemical effects of post-weaning social isolation in rodents-relevance to developmental neuropsychiatric disorders. Neuroscience & Biobehavioral Reviews. 32, 1087-1102.
- Fontaine, D.A., Davis, D.B. (2016). Attention to Background Strain Is Essential for Metabolic Research: C57BL/6 and the International Knockout Mouse Consortium. Diabetes. 65, 25-33.
- Johnson, M.S., Thomson, S.C., Speakman, J.R. (2001). Limits to sustained energy intake. III. Effects of concurrent pregnancy and lactation in Mus musculus. Journal of Experimental Biology. 204, 1947-1956.
- Lerch, S., Brandwein, C., Dormann, C., Gass, P., Chourbaji, S., (2015). Bred to breed?! Implications of continuous mating on the emotional status of mouse offspring. Behavioural Brain Research. 279, 155-165.
- Lloyd, S., (1983). Effect of pregnancy and lactation upon infection. Veterinary Immunology and Immunopathology. 4, 153-176.
- Mantalenakis, S.J., Ketchel, M.M., (1966). Frequency and extent of delayed implantation in lactating rats and mice. Journal of Reproduction and Fertility. 12, 391-394.
- Mccarthy, J.C., (1965). Effects of Concurrent Lactation of Litter Size and Prenatal Mortality in an Inbred Strain of Mice. Journal of Reproduction and Fertility. 9, 29-39.
- Norris, M. L., Adams, C.E., (1981). Concurrent lactation and reproductive performance in CFLP mice mated post partum. Laboratory Animals. 15, 273-275.
- Richmond, J., (2000). The 3Rs - Past, present and future. Scandinavian Journal of Laboratory Animal Science. 27. 84-92.
- Sayler, A., Salmon, M., (1969). Communal nursing in mice: influence of multiple mothers on the growth of the young. Science. 164, 1309-1310.
- Simon, M.M., Greenaway, S., White, J.K., Fuchs, H., Gailus-Durner, V., Wells, S., Sorg, T., Wong, K., Bedu, E., Cartwright, E.J., Dacquin, R., Djebali, S., Estabel, J., Graw, J., Ingham, N.J., Jackson, I.J., Lengeling, A., Mandillo, S., Marvel, J., Meziane, H., Preitner, F., Puk, O., Roux, M., Adams, D.J., Atkins, S., Ayadi, A., Becker, L., Blake, A., Brooker, D., Cater, H., Champy, M.F., Combe, R., Danecek, P., di Fenza, A., Gates, H., Gerdin, A. K., Golini, E., HancocK, J.M., Hans, W., Holter, S. M., Hough, T., Jurdic, P., Keane, T.M., Morgan, H., Muller, W., Neff, F., Nicholson, G., Pasche, B., Roberson, L.A., Rozman, J., Sanderson, M., Santos, L., Selloum, M., Shannon, C., Southwell, A., Tocchini-Valentini, G.P., Vancollie, V.E., Westerberg, H., Wurst, W., Zi, M., Yalcin, B., Ramirez-Solis, R., Steel, K.P., Mallon, A.M., De Angelis, M.H., Herault, Y., Brown, S.D., (2013). A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biology. 14, R82.
- Srivastava, A., Morgan, A.P., Najarian, M.L., Sarsani, V.K., Sigmon, J.S., Shorter, J.R., Kashfeen, A., McMullan, R.C., Williams, L.H., Giusti-Rodriguez, P., Ferris, M.T., Sullivan, P., Hock, P., Miller, D.R., Bell, T.A., McMillan, L., Churchill, G.A. & De Villena, F.P., (2017). Genomes of the Mouse Collaborative Cross. Genetics. 206, 537-556.
- Valzelli, L., (1973). The «isolation syndrome» in mice. Psychopharmacologia. 31, 305-320.
- van Loo, P.L., van Zutphen, L.F., Baumans, V., (2003). Male management: Coping with aggression problems in male laboratory mice. Laboratory Animals. 37, 300-313.
- Varty, G.B., Powell, S.B., Lehmann-Masten, V., Buell, M.R. & Geyer, M.A. (2006). Isolation rearing of mice induces deficits in prepulse inhibition of the startle response. Behavioural Brain Research. 169, 162-167.